Microarray Data Analysis
R scripts for analysis of microarray data.
We used microarray data to compare the expression of two different sets of genes from different cells maintained in particular conditions.
Outputs
Example
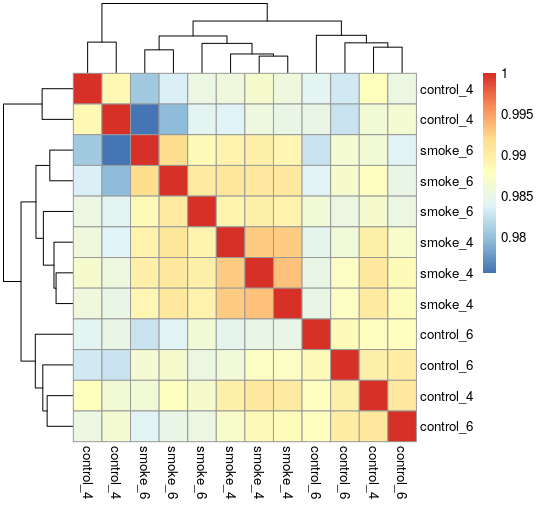
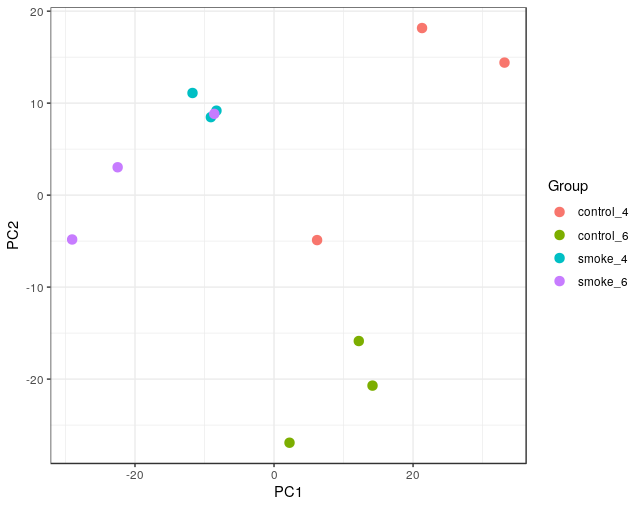
We used GEO dataset GSE13601 as an example. In this dataset there are expression data from cigarette smoke-treated mice at 4 and 6 months of age.
limma, Biobase, GEOquery, pheatmap, ggplot2, plyr, pheatmap
We used microarray data to compare the expression of two different sets of genes from different cells maintained in particular conditions.
Outputs
- Correlation matrix between samples
- Principal component analysis results
- List of the up-regulated genes
Example
We used GEO dataset GSE13601 as an example. In this dataset there are expression data from cigarette smoke-treated mice at 4 and 6 months of age.
- Subjects goups: control_4: Control mice at 4 months of age
- Correlation matrix between samples
- Principal component analysis results
- List of the up-regulated genes upregulated.txt
control_6: Control mice at 4 months of age
smoke_4: Cigarette smoke-treated mice at 4 months of age
smoke_6: Cigarette smoke-treated mice at 6 months of age
limma, Biobase, GEOquery, pheatmap, ggplot2, plyr, pheatmap